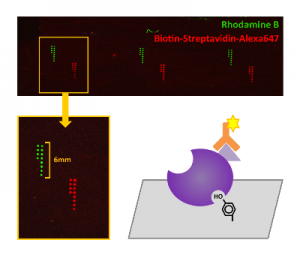
 Metabolic engineering efforts can be hampered and reduced to trial-and-error by complications arising from uncharacterized interactions within the host organism. These efforts would therefore benefit greatly from a systems-level understanding of metabolism. Systematic characterization strategies have been successfully applied at the genomic, transcriptomic, and proteomic levels, but downstream regulatory interactions are comparatively underexplored. Allosteric regulation is a common phenomenon by which metabolites bind to and alter the activity of proteins. Evidence suggests that current knowledge of allosteric interactions comprises only a small fraction of all existing cases. A broad understanding of allostery in Saccharomyces cerevisiae would therefore greatly accelerate efforts to create yeast strains that produce medically and economically important compounds, and a systematic methodology for discovery and characterization is necessary to obtain this understanding. Small molecule microarrays of yeast metabolites will provide a high-throughput platform for qualitatively identifying protein-metabolite binding pairs by probing the arrays with tagged, known proteins. Additionally, in vitro assays coupled with gas chromatography analysis will be carried out in parallel to probe interactions in parts of the metabolome that microarrays may not effectively capture. The combined information from these complementary approaches will facilitate the eventual construction of a systems-level mathematical model of S. cerevisiae metabolism.
Metabolic engineering efforts can be hampered and reduced to trial-and-error by complications arising from uncharacterized interactions within the host organism. These efforts would therefore benefit greatly from a systems-level understanding of metabolism. Systematic characterization strategies have been successfully applied at the genomic, transcriptomic, and proteomic levels, but downstream regulatory interactions are comparatively underexplored. Allosteric regulation is a common phenomenon by which metabolites bind to and alter the activity of proteins. Evidence suggests that current knowledge of allosteric interactions comprises only a small fraction of all existing cases. A broad understanding of allostery in Saccharomyces cerevisiae would therefore greatly accelerate efforts to create yeast strains that produce medically and economically important compounds, and a systematic methodology for discovery and characterization is necessary to obtain this understanding. Small molecule microarrays of yeast metabolites will provide a high-throughput platform for qualitatively identifying protein-metabolite binding pairs by probing the arrays with tagged, known proteins. Additionally, in vitro assays coupled with gas chromatography analysis will be carried out in parallel to probe interactions in parts of the metabolome that microarrays may not effectively capture. The combined information from these complementary approaches will facilitate the eventual construction of a systems-level mathematical model of S. cerevisiae metabolism.