New AI Tool Identifies Better Antibody Therapies
New AI Tool Identifies Better Antibody Therapies
From sending cancer into remission to alleviating Covid-19 symptoms, immunotherapy can provide revolutionary disease treatments. Immunotherapies use antibodies — proteins that bind to cell markers called antigens — to target and eliminate the antigen. But despite how effective immunotherapy can be, it isn’t widely used because finding the right antibodies to develop treatments is challenging, time-consuming work.
Georgia Tech researchers are making this process a little easier, though. Their new tool, AF2Complex, used deep learning to predict which antibodies could bind to Covid-19’s infamous spike protein. The researchers created input data for the deep-learning model using sequences of known antigen binders. This method correctly predicted 90% of the best antibodies in one test with 1,000 antibodies and was recently published in Proceedings of the National Academy of Sciences. Treating Covid-19 is just the start of its potential.
 Trial Begins For ‘Probabilistic Approach’ To Diagnosing Ovarian Cancer
Trial Begins For ‘Probabilistic Approach’ To Diagnosing Ovarian Cancer
Scientists in the Georgia Tech Integrated Cancer Research Center (ICRC) are proposing a new, more realistic approach to diagnosing cancer that provides a probabilistic statement about the likelihood of developing it, much like how cholesterol tests are used to assess heart disease risk. Probabilities are based on an individual’s metabolic profile and the first population group to be targeted are women at high risk for ovarian cancer, according to John McDonald, Ph.D., professor emeritus in the school of biological sciences at Georgia Institute of Technology and founding director of the ICRC.
*Also featured on Clinical Research News.
Researchers Leverage AI to Develop Early Diagnostic Test for Ovarian Cancer
For over three decades, a highly accurate early diagnostic test for ovarian cancer has eluded physicians. Now, scientists in the Georgia Tech Integrated Cancer Research Center (ICRC) have combined machine learning with information on blood metabolites to develop a new test able to detect ovarian cancer with 93 percent accuracy among samples from the team’s study group.
*Also featured on the New York Post, Atlanta New First, Medical Xpress, News Medical Life Sciences, Medscape, AJMC, ScienceDaily, Life Technology, Omics Tutorials, Oncology News Australia, Newswise and Eurekalert.
 The University System of Georgia (USG) Board of Regents has affirmed the renewal of Jeffrey Skolnick as a Regents’ Professor!
The University System of Georgia (USG) Board of Regents has affirmed the renewal of Jeffrey Skolnick as a Regents’ Professor!
Regents’ distinctions may be granted for a period of three years by the Board of Regents (BOR) to outstanding faculty members from Georgia Tech, Augusta University, Georgia State University, the University of Georgia, and, in special circumstances, other USG institutions. A Regents’ professor, researcher, or entrepreneur distinction is awarded only after unanimous recommendation from the president of the recipient’s university, their chief academic officer and dean, as well as three additional members of the faculty who are named by the university president.
 Mu Gao wins the 2023 Outstanding Sr. Research Faculty Award!
Mu Gao wins the 2023 Outstanding Sr. Research Faculty Award!
Mu Gao was awarded with the 2023 Outstanding Sr. Research Faculty Award by the Georgia Tech College of Sciences at the Spring Science Celebration, Celebrating Excellence in Research, Teaching, and Community hosted by Susan Lozier, Dean of the College of Sciences and Betsy Middleton and john Clark Sutherland Chair.
 AF2Complex ‘Computational Microscope’ Predicts Protein Interactions, Potential Paths to New Antibiotics
AF2Complex ‘Computational Microscope’ Predicts Protein Interactions, Potential Paths to New Antibiotics
Though it is a cornerstone of virtually every process that occurs in living organisms, the proper folding and transport of biological proteins is a notoriously difficult and time-consuming process to experimentally study.
 Computing function from form
Computing function from form
Georgia Tech and Oak Ridge National Laboratory researchers extend AI’s prediction prowess to complex protein structures.
Over the past two years, artificial intelligence has shown it can predict what many cellular components look like. For instance, the AlphaFold deep-learning tool developed by Google sister company DeepMind has decoded how nearly every amino acid sequence folds into defined shapes.
AF2Complex: Researchers Leverage Deep Learning to Predict Physical Interactions of Protein Complexes
A computational tool developed to predict the structure of protein complexes – the molecular machinery that makes life possible – is lending new insights into the biomolecular mechanisms of their function.
Scientists use Summit supercomputer, deep learning to predict protein functions at genome scale
(olfc.ornl.gov, January 10, 2022) A team of scientists led by the Department of Energy’s Oak Ridge National Laboratory and the Georgia Institute of Technology is using supercomputing and revolutionary deep learning tools to predict the structures and roles of thousands of proteins with unknown functions.

Georgia Tech researchers making breakthroughs treating COVID-19
(WSB-TV Channel 2 Action, November 16, 2021) News coverage of MOATAI-VIR.
 Emory, Georgia Tech researchers develop AI-tool to predict COVID-19 symptoms
Emory, Georgia Tech researchers develop AI-tool to predict COVID-19 symptoms
(emorywheel.com, November 12, 2021) Researchers at Emory and the Georgia Institute of Technology (Georgia Tech) published a paper on Oct. 21 detailing an artificial intelligence-based tool that is able to predict COVID-19 symptoms and suggest specific FDA-approved drugs.

AI Tool Pairs Protein Pathways with Clinical Side Effects, Patient Comorbidities to Suggest Targeted Covid-19 Treatments
(cos.gatech.edu, October 21, 2021)The symptoms and side effects of Covid-19 are scattered across a diagnostic spectrum. Some patients are asymptomatic or experience a mild immune response, while others report significant long-term illnesses, lasting complications, or suffer fatal outcomes.
Three researchers from the Georgia Institute of Technology and one from Emory University are trying to help clinicians sort through these factors and spectrum of patient outcomes by equipping healthcare professionals with a new “decision prioritization tool.” PDF
 Bartosz Ilkowski Promoted to Principal Research Technologist
Bartosz Ilkowski Promoted to Principal Research Technologist
(cos.gatech.edu, July 15, 2021) Five Researchers Gain New Titles in the College of Sciences – School of Biological Sciences, School of Chemistry and Biochemistry scientists rewarded for work, support, and service. In the School of Biological Sciences, the following promotions have occurred:
Bartosz Ilkowski — principal research technologist (formerly senior research technologist)
Kuntal Mukerjee — senior research scientist (formerly research scientist II)
Brandy Olmer — research associate II (formerly research associate)
 ASPIRE-ing To Find Fast Solutions to the Opioid Health Crisis
ASPIRE-ing To Find Fast Solutions to the Opioid Health Crisis
(cos.gatech.edu, May 27, 2021) School of Biological Sciences’ Jeffrey Skolnick and Hongyi Zhou are part of an award-winning NIH effort to create innovative, AI-powered platforms for discovering new pain management drugs — and identify immediate solutions
Congratulations 2020 Outstanding Bioinformatics Students
(bioinformatics.gatech.edu, August 27, 2020) Congratulations to Alli Gombolay (PhD), Courtney Astore (MS), Sonali Gupta (MS), and Aaron Pfennig (MS) who were recognized as the 2020 Outstanding Students in Bioinformatics at Georgia Tech. Each student received a monetary award funded by the J. Leland Jackson Endowed Fellowships Fund in Bioinformatics. Students were selected for demonstrated excellence in their academic studies and their bioinformatics research.
 Stay at Home Summer: Courtney Astore Balances a Packed Schedule with Comedy, Community Connections
Stay at Home Summer: Courtney Astore Balances a Packed Schedule with Comedy, Community Connections
(cos.gatech.edu, July 28, 2020) Courtney Astore is a second-year Ph.D. student in the Bioinformatics program. While her summer days are packed with virtual classes and research activities, Astore shares that she’s also set aside time for exercise, time with family, and leisure activities like reading.
 Courtney Astore Awarded 2020 HIMSS Scholarship
Courtney Astore Awarded 2020 HIMSS Scholarship
(bioinformatics.gatech.edu, June 4, 2020) Congratulations to Bioinformatics doctoral student, Courtney Astore, who was recently selected for a 2020 Healthcare Information and Management Systems Society (HIMSS) Scholarship. Courtney will be awarded $3000 for this honor. Students were evaluated on the basis of their academic achievements, engagement with health information technology, leadership, and vision for their future careers.
 Jeffrey Skolnick Receives Regents Recognition
Jeffrey Skolnick Receives Regents Recognition
(news.gatech.edu, May 7, 2020)Seven Georgia Tech Faculty Members Receive Regents Recognition – The University System of Georgia (USG) Board of Regents (BOR) appointed seven Georgia Tech faculty members Regents Professors, the highest academic recognition bestowed by the USG. The seven Regents Professors are:
- Marilyn Brown, Brook Byers Professor in Sustainable Manufacturing in the School of Public Policy
- Suresh Sitaraman, Morris M. Bryan Jr. Professor in Mechanical Engineering in the George W. Woodruff School of Mechanical Engineering
- Jeffrey Skolnick, Mary and Maisie Gibson Chair in Computational Systems Biology and GRA Eminent Scholar in the School of Biological Sciences
- Prasad Tetali, professor in the School of Mathematics and the School of Computer Science
- Vigor Yang, professor in the Daniel Guggenheim School of Aerospace Engineering
- Lisa Yaszek, professor in the School of Literature, Media, and Communication
- Ellen Zegura, Stephen Fleming Chair in the School of Computer Science
Origin of Life’s Handedness and Protein Biochemistry
(cos.gatech.edu, December 11, 2019) Examine your hands. The right is a mirror image of the left. They look very similar, but you know they’re not when you try to put your left hand inside a right glove.
The molecules of life have a similar handedness. Proteins for example are like your left hand, made up of amino acids that are all left-handed. This phenomenon is called chirality. How chiral systems emerged is one of the key questions of origins-of-life research.
 On the possible origin of protein homochirality, structure, and biochemical function
On the possible origin of protein homochirality, structure, and biochemical function
(PNAS, December 10, 2019) Living systems contain mainly chiral macromolecules, including proteins. How L-chiral proteins emerged from demi-chiral mixtures is unknown. Our simulations show that, compared to contemporary proteins, demi-chiral proteins have shorter regular secondary structures due to fewer internal hydrogen bonds, but similar global folds and small molecule binding sites. Demi-chiral proteins contain L-chiral substructures matching native active site geometries. Among the most frequently generated enzymes with native active site residues are ancient functions associated with metabolism and replication. This suggests that demi-chiral proteins could engage in early metabolism, creating the feedback loop for transcription and cell formation partly responsible for life’s emergence.
 Jeffrey Skolnick, Mu Gao, and Hongyi Zhou win the NCATS ASPIRE Design Challenge 3: Predictive Algorithms for Translational Innovation in Pain, Opioid Use Disorder and Overdose
Jeffrey Skolnick, Mu Gao, and Hongyi Zhou win the NCATS ASPIRE Design Challenge 3: Predictive Algorithms for Translational Innovation in Pain, Opioid Use Disorder and Overdose
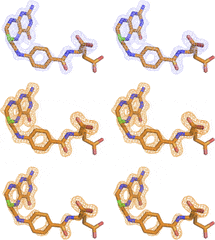 Time-resolved x-ray crystallography capture of a slow reaction tetrahydrofolate intermediate
Time-resolved x-ray crystallography capture of a slow reaction tetrahydrofolate intermediate
Structural Dynamics – Editor’s Pick
 Jeffrey Skolnick: 2018 Sigma Xi Sustained Research Award – Applying computational systems biology to improve human health
Jeffrey Skolnick: 2018 Sigma Xi Sustained Research Award – Applying computational systems biology to improve human health
(cos.gatech.edu, April 5, 2018) Georgia Tech has named Jeffrey Skolnick the recipient of the 2018 Sigma Xi Sustained Research Award. The award recognizes Skolnick’s exceptional sustained imagination and productivity in the fields of systems biology, computational biology, bioinformatics, cancer metabolomics, protein structure prediction and evolution, drug design, and simulations of cellular processes.
Drug Design Strategy Boosts the Odds Against Resistance Development
(news.gatech.edu, June 14, 2017) A new rational drug design technique that uses a powerful computer algorithm to identify molecules that target different receptor sites on key cellular proteins could provide a new weapon in the battle against antibiotic resistance, potentially tipping the odds against the bugs.
 Rattling DNA Hustles Transcribers to Targets
Rattling DNA Hustles Transcribers to Targets
(news.gatech.edu, June 12, 2017) New simulations of DNA as a transport conduit could shatter the way scientists have thought about how large molecules called transcription factors diffuse on their way to carry out genetic missions, according to a study by researchers at the Georgia Institute of Technology. The simulations add important brush strokes to our picture of the elusive inner mechanics of cells.
Billion-dollar project would synthesize hundreds of thousands of molecules in search of new medicines
(Science News, April 19, 2017) Two years ago, Martin Burke estimated that assembling 75% of natural products with his machine would take some 5000 different building blocks, compared with just four for DNA—a challenging number for chemical suppliers to make and stock. But now, Burke told the ACS meeting, the problem looks more manageable. His lab recently teamed up with that of Jeffrey Skolnick, a computational biologist at the Georgia Institute of Technology in Atlanta.