for samples in organic solvents:
- First run 1D PROTON, and find proper sw, o1, and p1 by pulsecal.
- Run 2D DOSY with dstebpgp3s sequence — with convection compensation,
- Set d1 (3s here) + aq (0.75s here) >= 3*T1, 5*T1 is better;
- input the proper sw, p1, o1 values from step 1; and check those gradient pulses (gpz6,7,8,9) are set correctly,
- edp, make: lb=1 or 2, absf1=1000, 1000; absf2= -1000, -1000; absg = 5,
- keep p30 =1000us (default on B700, can be 500us to 3000us—RT probe, ≤ 2000us on cryoprobe) and d20 = 60ms (default on B700; d20=50ms is ok for mw=350 Da molecule; can be: 10ms to 70ms), set ns= 16 and type dosy and run with gpz6=3% to 3% with 1 point and choose q, run rga, keep rg, and run dosy again with gpz6=3% to 95% , and 7 (or 9) points, choose q, use the same previous rg value.
- process data with T1T2 Application, and see how the decay graph looks like:
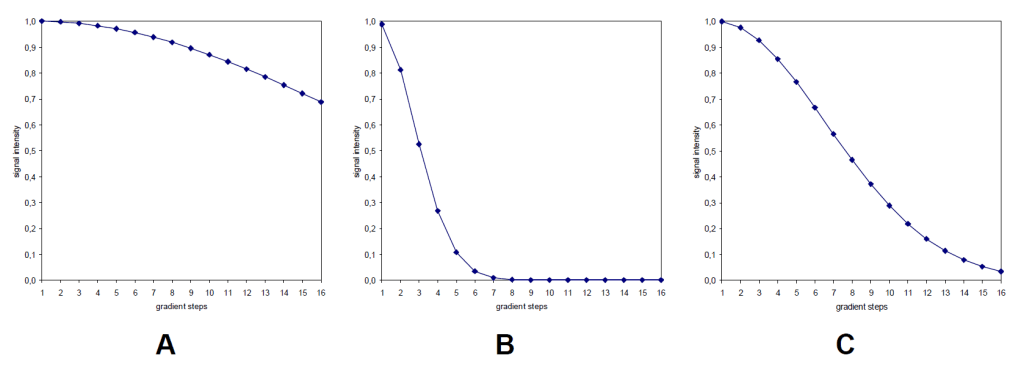
- if A, increase d20; if B, decrease d20; if C, d20 is proper.
- Re-run dosy with ns=16 or 32, gpz6 from 3% to 95%, 7 points, q; and process with T1T2 Applications again.
Note:
- dstebpgp3s sequence is mostly used at all Temp., dstebpgp3s1d sequence is ready too,
- make sure gradient duty cycle p30/(d1 + aq) < 5%,
- ledbpgp2s — (w/o convection compensation; 1D sequence: ledbpgp2s1d), is also commonly used.
for samples in 90%H2O/10%D2O, need water suppression:
- stebpgp1s19: — without cc and led; with watergate3-9-19; since the peptide/protein’s NH exchanges with water, the measured D from NH region peaks will be the average value between time spent on NH and H2O, so use non-exchangeable protons peaks to determine D.
- ledbpgppr2d: — water suppression with presaturation, without cc.
Hongwei edited on 4/27/2023

